K-Means¶
K-means is one of the simplest and most popular unsupervised machine learning algorithms. The goal of K-means is to group similar data points together and discover underlying patterns. To achieve this objective, K-means looks for a fixed number (k) of clusters in a dataset.
A cluster refers to a collection of data points aggregated together because of certain similarities. The 'K' in K-means represents the number of clusters the algorithm tries to identify in the dataset.
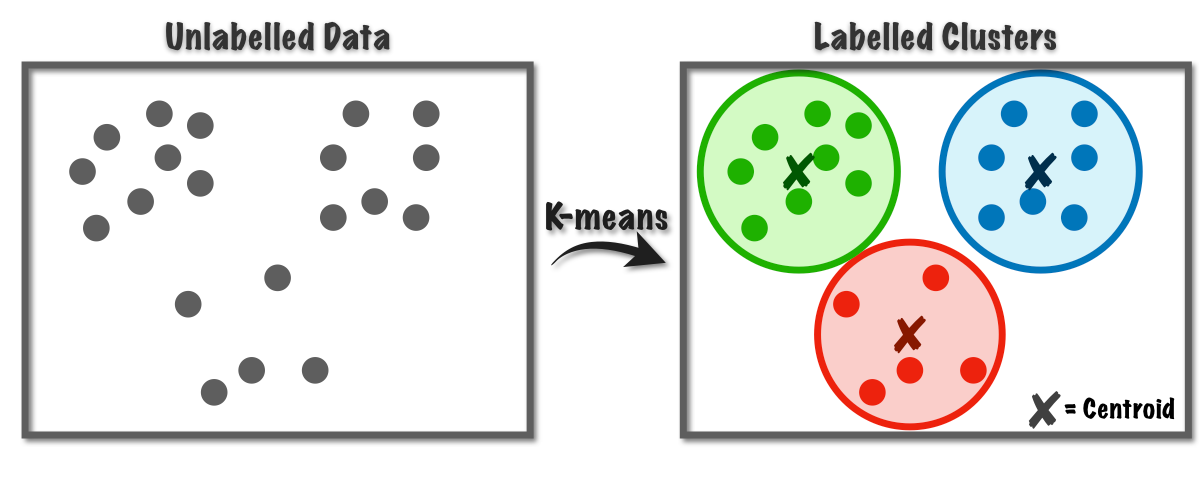
Algorithm idea¶
The K-means algorithm works iteratively to assign each data point to one of $k$ groups based on the features provided.
Steps:
- Initialization: Randomly initialize $k$ centroids.
- Assignment: Assign each data point to the nearest centroid. This forms the clusters.
- Update: Calculate the new means (centroids) of each cluster.
- Repeat: Repeat steps 2 and 3 until the centroids do not change significantly.
Mathematically, the objective of the K-means clustering is to minimize the within-cluster sum of squares (WCSS). It can be represented as : $$\mathrm{WCSS}=\textstyle{\sum_{i=1}^{k}}\sum_{x\in C_{i}}|x-\mu_{i}||^{2}$$ where $C_{i}$ is the $i$th cluster and $\mu_{i}$ is the centroid of the $i$th cluster and x is a data point in cluster $C_{i}$.
This is an animation of the K-means algorithm in action. Notice how the centroids (black cross) move with each iteration 🤓
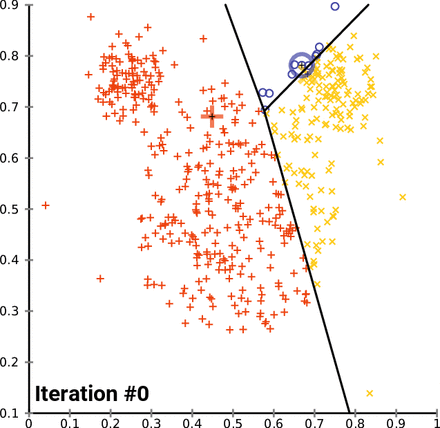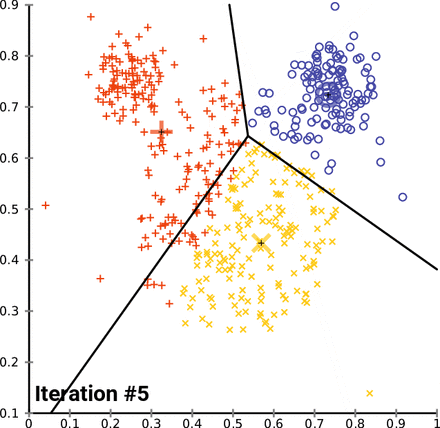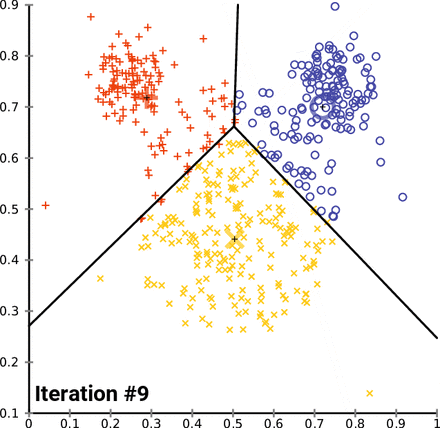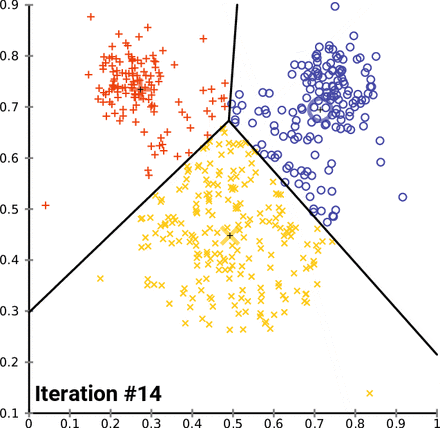
Example in scikit-learn¶
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets import load_iris
iris = load_iris()
X = iris.data
y = iris.target
from sklearn.cluster import KMeans
# Applying KMeans to the Mall dataset
kmeans = KMeans(n_clusters= 3, init= "k-means++", max_iter=300, n_init = 10, random_state=0)
y_kmeans = kmeans.fit_predict(X)
# Visualizing the clusters and centroids
plt.scatter(X[y == 0,2], X[y == 0,3], s = 100, c = 'red')
plt.scatter(X[y == 1,2], X[y == 1,3], s = 100, c = 'green')
plt.scatter(X[y == 2,2], X[y == 2,3], s = 100, c = 'blue')
plt.scatter(kmeans.cluster_centers_[:,2], kmeans.cluster_centers_[:,3], s = 300, c = 'yellow', label = 'centroids')
plt.title('clusters of iris')
plt.xlabel('petal width')
plt.ylabel('Sepal length')
plt.legend()
plt.show()
# Visualisation with dimension reduction
from sklearn.decomposition import PCA
reduced_data = PCA(n_components=2).fit_transform(X)
kmeans = KMeans(init='k-means++', n_clusters=3, n_init=10)
kmeans.fit(reduced_data)
KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=1, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
plt.scatter(reduced_data[y == 0,0], reduced_data[y == 0,1], s = 100, c = 'red')
plt.scatter(reduced_data[y == 1,0], reduced_data[y == 1,1], s = 100, c = 'green')
plt.scatter(reduced_data[y == 2,0], reduced_data[y == 2,1], s = 100, c = 'blue')
plt.scatter(kmeans.cluster_centers_[:,0], kmeans.cluster_centers_[:,1], s = 300, c = 'yellow')
<matplotlib.collections.PathCollection at 0x1a1e400748>
Choosing $k$¶
One of the challenges with K-means is that we need to specify the number of clusters (k) before running the algorithm 😅. Several methods can help determine the optimal number of clusters:
- Elbow Method: Plot the number of clusters against the WCSS. The point where the reduction in WCSS begins to slow down (elbow point) is considered a good estimate for 'k'.
- Silhouette Method: Measures the quality of clusters. A higher silhouette score indicates better-defined clusters.
Advantages and Disadvantages¶
Advantages¶
- Simple and easy to implement.
- Efficient in terms of computational cost.
- Scales well to large datasets.
Disadvantages¶
- Assumes clusters to be spherical and equally sized.
- Sensitive to the initial placement of centroids.
- Can get stuck in local optima. Often, multiple runs with different initializations are required.
- Requires the number of clusters to be specified beforehand.
Variations and Improvements¶
- K-means++: Improves the initialization of the centroids to provide better convergence.
- Mini Batch K-means: Uses a random sample of data points in each iteration for updating the clusters, making it faster.
- Bisecting K-means: A hierarchical approach where clusters are split iteratively.
# Using the Elbow method to find the optimal number K of clusters
from sklearn.cluster import KMeans
wcss = []
for i in range (1,11):
kmeans = KMeans(n_clusters= i, init = "k-means++", max_iter = 300, n_init = 10, random_state = 0)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
plt.plot(range(1,11), wcss)
plt.title('Elbow Method')
plt.xlabel('Number of Clusters')
plt.ylabel('WCSS')
plt.show()
Metrics for evaluating clustering performance¶
There are several metrics for evaluating clustering performance. We will use the following metrics in this notebook :
- Homogeneity: A clustering result satisfies homogeneity if all of its clusters contain only data points which are members of a single class.
- Completeness: A clustering result satisfies completeness if all the data points that are members of a given class are elements of the same cluster.
- V-measure: The V-measure is the harmonic mean between homogeneity and completeness.
- Silhouette Coefficient: The Silhouette Coefficient is calculated using the mean intra-cluster distance (a) and the mean nearest-cluster distance (b) for each sample. The Silhouette Coefficient for a sample is (b - a) / max(a, b). To clarify, b is the distance between a sample and the nearest cluster that the sample is not a part of. Note that Silhouette Coefficient is only defined if number of labels is 2 <= n_labels <= n_samples - 1.
Implementation of Silhouette Coefficient in scikit-learn¶
from sklearn.datasets import make_blobs
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_samples, silhouette_score
import matplotlib.cm as cm
for n_clusters in range(2,4):
# Create a subplot with 1 row and 1 columns
fig, ax1 = plt.subplots(1, 1)
fig.set_size_inches(18, 7)
# The 1st subplot is the silhouette plot
# The silhouette coefficient can range from -1, 1 but in this example all
# lie within [-0.1, 1]
ax1.set_xlim([-0.1, 1])
# The (n_clusters+1)*10 is for inserting blank space between silhouette
# plots of individual clusters, to demarcate them clearly.
ax1.set_ylim([0, len(X) + (n_clusters + 1) * 10])
# Initialize the clusterer with n_clusters value and a random generator
# seed of 10 for reproducibility.
clusterer = KMeans(n_clusters=n_clusters, random_state=10)
cluster_labels = clusterer.fit_predict(X)
# The silhouette_score gives the average value for all the samples.
# This gives a perspective into the density and separation of the formed
# clusters
silhouette_avg = silhouette_score(X, cluster_labels)
print("For n_clusters =", n_clusters,
"The average silhouette_score is :", silhouette_avg)
# Compute the silhouette scores for each sample
sample_silhouette_values = silhouette_samples(X, cluster_labels)
y_lower = 10
for i in range(n_clusters):
# Aggregate the silhouette scores for samples belonging to
# cluster i, and sort them
ith_cluster_silhouette_values = \
sample_silhouette_values[cluster_labels == i]
ith_cluster_silhouette_values.sort()
size_cluster_i = ith_cluster_silhouette_values.shape[0]
y_upper = y_lower + size_cluster_i
color = cm.nipy_spectral(float(i) / n_clusters)
ax1.fill_betweenx(np.arange(y_lower, y_upper),
0, ith_cluster_silhouette_values,
facecolor=color, edgecolor=color, alpha=0.7)
# Label the silhouette plots with their cluster numbers at the middle
ax1.text(-0.05, y_lower + 0.5 * size_cluster_i, str(i))
# Compute the new y_lower for next plot
y_lower = y_upper + 10 # 10 for the 0 samples
ax1.set_title("The silhouette plot for the various clusters.")
ax1.set_xlabel("The silhouette coefficient values")
ax1.set_ylabel("Cluster label")
# The vertical line for average silhouette score of all the values
ax1.axvline(x=silhouette_avg, color="red", linestyle="--")
ax1.set_yticks([]) # Clear the yaxis labels / ticks
ax1.set_xticks([-0.1, 0, 0.2, 0.4, 0.6, 0.8, 1])
plt.show()
For n_clusters = 2 The average silhouette_score is : 0.6808136202713507 For n_clusters = 3 The average silhouette_score is : 0.5525919445213676
Conclusion¶
K-means is a foundational algorithm for cluster analysis in data mining. Despite its simplicity, K-means can achieve robust clustering results on a wide range of datasets, especially when the shape and number of clusters are appropriate for the algorithm's assumptions. It's also computationally efficient and easy to implement. However, K-means has several drawbacks, such as the need to specify the number of clusters beforehand and the sensitivity to the initial placement of centroids.